Citing CLIME
 Y. Li, S.E. Calvo, R. Gutman, J.S. Liu, and V.K. Mootha (2014) Expansion of biological pathways based on evolutionary inference, Cell 158, 213–225. Pubmed: 24995987
Y. Li, S.E. Calvo, R. Gutman, J.S. Liu, and V.K. Mootha (2014) Expansion of biological pathways based on evolutionary inference, Cell 158, 213–225. Pubmed: 24995987
 Y. Li, S. Ning, S.E. Calvo, V.K. Mootha and J.S. Liu (2018) Bayesian hidden Markov tree models for clustering genes with shared evolutionary history, The Annals of Applied Statistics, in press. arXiv: 1808.06109
Y. Li, S. Ning, S.E. Calvo, V.K. Mootha and J.S. Liu (2018) Bayesian hidden Markov tree models for clustering genes with shared evolutionary history, The Annals of Applied Statistics, in press. arXiv: 1808.06109
Run CLIME
Downloads
CLIME software (including users' manual)
Pre-computed CLIME analyses of published gene sets
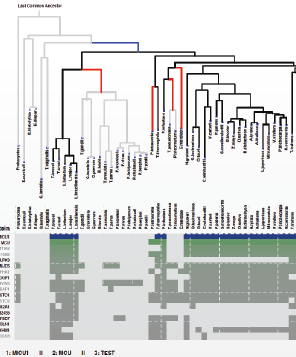
| TXT | Human complex I | |
| TXT | Human MICU1 | |
| TXT | Human ciliary genes | |
| TXT | Human mitochondrial genes | |
| TXT | All 5331 P. falciparum genes | |
| TXT | All 5013 C. merolae genes | |
| TXT | All 5882 S. cerevisiae genes | |
| 1025 human pathways from GO and KEGG | ||
Phylogenetic profile matrices for 10 model organisms across 138 eukaryotes (plus Prokaryote outgroup)
| hsa.matrix138.txt | H. sapiens phylogenetic matrix |
| mmu.matrix138.txt | M. musculus phylogenetic matrix |
| dme.matrix138.txt | D. melanogaster phylogenetic matrix |
| cel.matrix138.txt | C. elegans phylogenetic matrix |
| ncr.matrix138.txt | N. crassa phylogenetic matrix |
| sce.matrix138.txt | S. cerevisiae phylogenetic matrix |
| ath.matrix138.txt | A. thaliana phylogenetic matrix |
| cme.matrix138.txt | C. merolae phylogenetic matrix |
| pfa.matrix138.txt | P. falciparum phylogenetic matrix |
| tbr.matrix138.txt | T. brucei phylogenetic matrix |
| euk138.with_prok.nwk | Tree topology for 138 eukaryotes plus prokaryote outgroup, in Newick format |
Last updated 10/18/2022